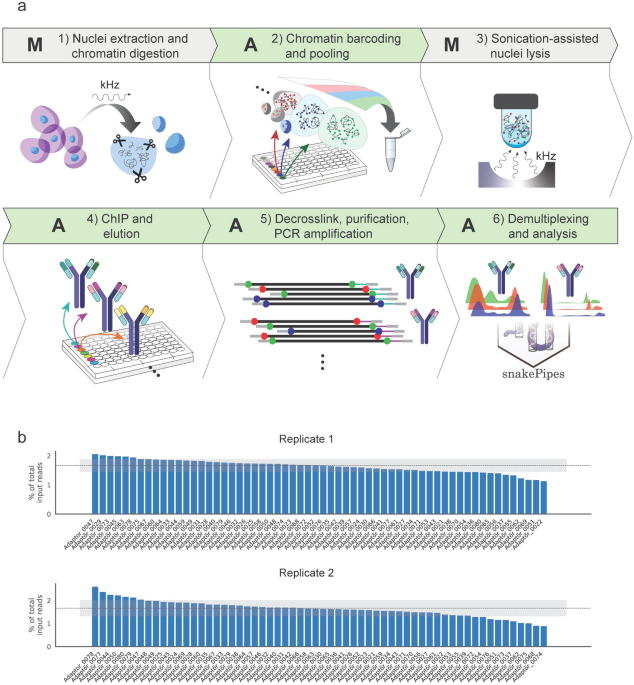

Consistently, 1% of sequencing reads from the Agencourt AMPure XP kit is smaller than 100 bp but nearly 5–10% of sequencing reads are smaller than 100 bp from column-based purification reagents (Additional file ). We tested whether purification reagents utilizing silica columns or SPRI may introduce any bias in ChIP-seq data. AMPure XP clean-ups. AMPure XP can be performed either manually or automated on a liquid handling system. Difference in time between manual and automation is indicated. NR = Not Recommended For use in manual or automated methods based on batch size or overall throughput. Agencourt AMPure XP Beads (Beckman Coulter, cat. A63880) are required and should be ordered separately. Please follow the manufacturer's instructions for storage and handling of the AMPure XP Reagent. ®GeneRead Adapters are fully compatible with Illumina instruments such as MiSeq, NextSeq® or HiSeq® instruments. The enrichment step is. AGENCOURT® RNACLEAN XP® Protocol 001298v001 Page 2 of 2 For questions regarding this protocol, call Technical Support at Agencourt 1-800-773-9186 Agencourt Bioscience Corporation, A Beckman Coulter Company 800-361-7780 978-867-2600 500 Cummings Center, Suite 2450 Beverly, Massachusetts 01915 www.agencourt.com.
I wanted to talk a little about the selection characteristics of Agencourt’s AMPure beads, a bead-reagent combination that purifies PCR reactions.
This stuff is incredible in terms of simplicity, efficiency, and high-throughput compatibility. I have a sneaking suspicion that AMPure, not unlike fire to Prometheus, was handed down from the gods to benefit humanity. You just dunk it into your sample, slosh it around, stick it to a magnet, wash, wash again, and elute in your favorite buffer. No muss, no fuss.
We were wondering, though, about its selection process. What size fragments are selected by the AMPure beads, specifically at which ratio of beads to sample? So, like diligent scientists, we rolled up the sleeves of our labcoats and… read the protocol.

The protocol recommends washing your sample in a 1.8:1 ratio of beads to sample, although it says that fragments less than 100bp will be omitted at this ratio, it doesn’t say which sized fragments will be selected. We found this remarkably helpful technical bulletin, which describes calibrating each batch of AMPure beads with various ratios of DNA ladder.
So I did our very own calibration with AMPure beads using Fermentas’s GeneRuler™ Low Range DNA Ladder (25-700 bp). I added 30ul ladder to various concentrations of AMPure beads according to Agencourt’s instructions.
(Actually, if you’re looking for good AMPure instructions, I recommend looking at Illumina’s TruSeq™ Sample Preparation Guide. Honestly, their instructions are more comprehensive than Agencourt’s, and easier to read.) After purifying each sample, I bookended the various AMPure:ladder ratios with 10ul non-purified ladder on a 2% TBE gel for easy comparison.
Beckman Ampure
Without any further ado, here are the results: